Microscoop®


Overview
Microscoop is a breakthrough platform for spatial proteomics, enabling researchers to map thousands of proteins inside single cells and subcellular compartments. By combining advanced microscopy with automated proteomics, Microscoop reveals the hidden complexity of biology — from cellular architecture to disease mechanisms.
Cancer & Disease Mechanism Research
Highlights
Explore cellular and subcellular interactions that shape health and disease like never before.
Unbiased discovery
Extract all proteomic information without antibodies or targeted panels across an entire sample.
High resolution
350 nm precision at sub-cellular scale.
Broad sample compatibility
Use with FFPE/fresh frozen tissue samples or fixed cells.
Identify targets faster
Discover novel biomarkers or therapeutic targets of disease-associated locations.
Superior sensitivity
Discover low copy number proteins from increased dynamic range due to subcellular protein isolation.
Reveal
Novel proteomes of organelles, cells and organisms.
Unmatched specificity
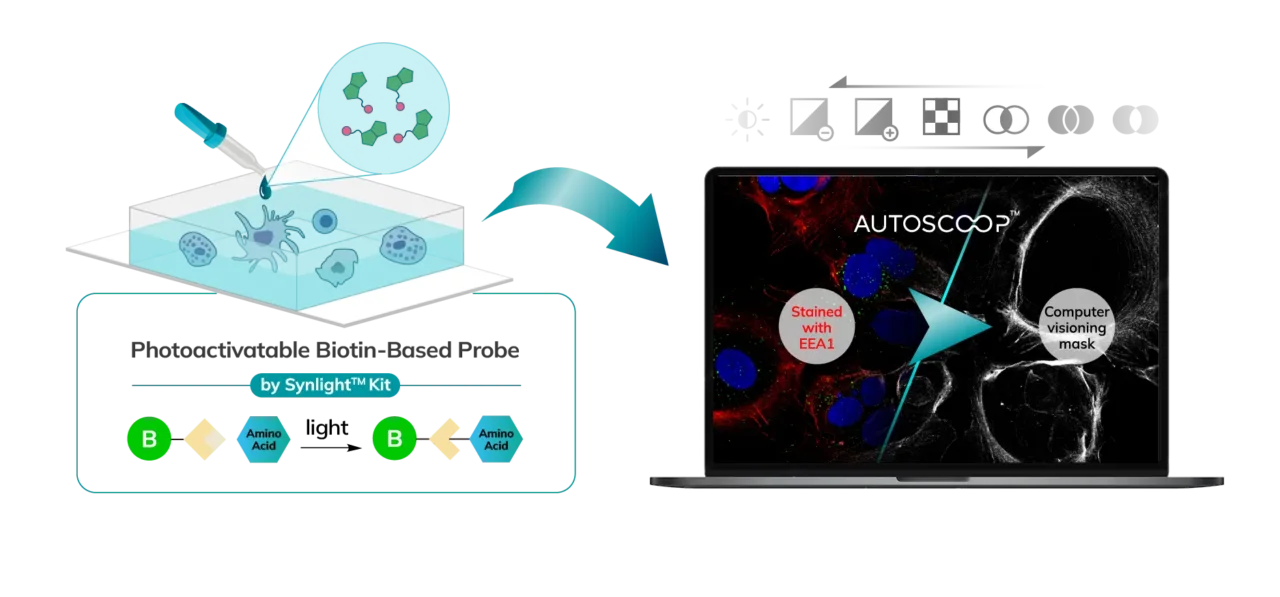
Two photon guided photo-biotinylation identifies only proteins in regions of interest.
Maximize the value and impact of your mass spec data
Traditional proteomic technology often relies on predefined panels or antibody-based enrichment,
which introduces bias—only known or expected proteins are detected.
But Syncell’s Microscoop® labels and analyzes all proteins in a specific region of interest
without pre-selection, allowing mass spec to capture low-abundance or rare proteoforms, and reveal
otherwise unknown protein constituents with higher dynamic range and specificity than laser
microdissection or proximity labeling methods.
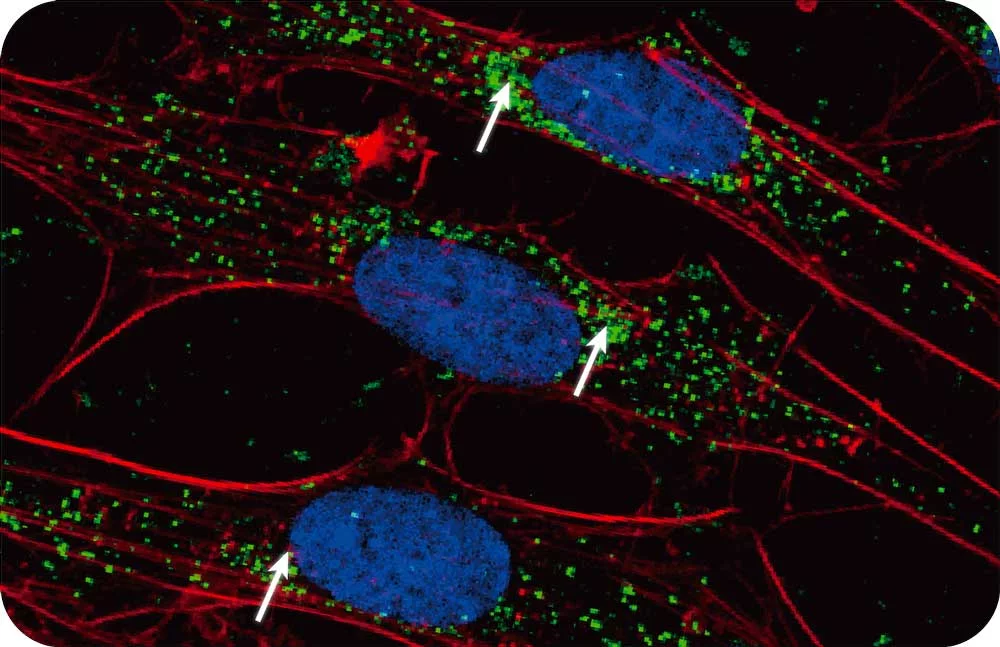
Mass spec alone doesn’t provide spatial information, but combining it with Syncell’s microscopy-guided photo-biotinylation technology enables researchers to link spatial location with the entire local proteome, allowing you to:
- Understand the entire proteomic makeup of key organelles (e.g., cilia, nuclei, tumor microenvironments).
- Understand the location–function relationships of proteins.
- Identify disease-related signatures based on tissue architecture to pinpoint exact proteomic drivers of progression.
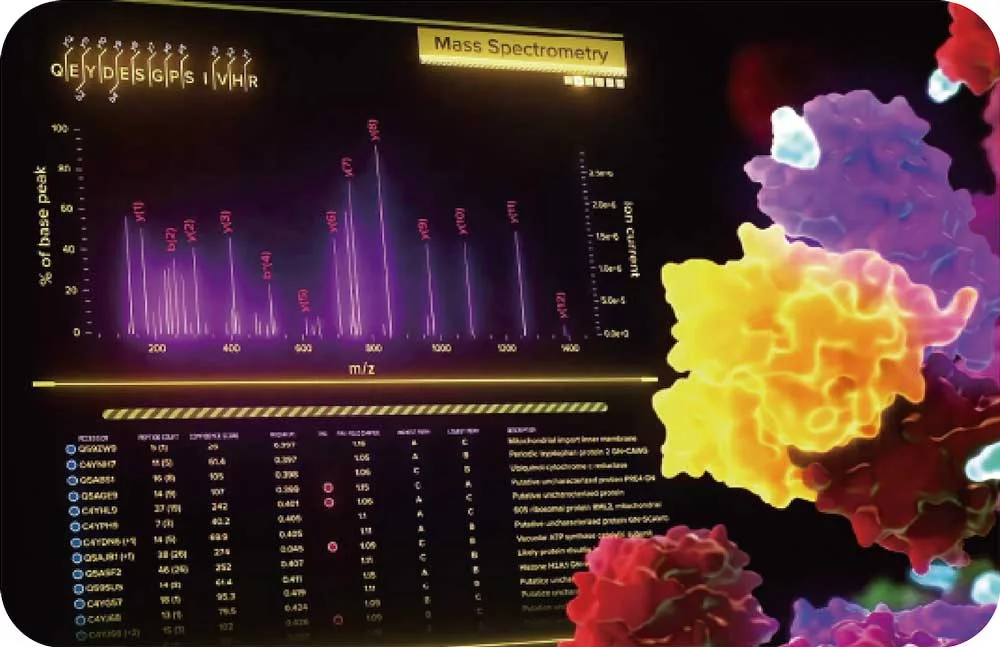
By restricting proteomic analysis to defined cellular or subcellular regions, our proprietary opto-proteomic platform improves:
- Signal-to-noise in complex tissue samples
- Dynamic range and detection of low-expressing proteins in LC/MS data
- Single-region proteomic profiling

Microscoop® works with FFPE and fresh frozen tissue, preserving the integrity of archived clinical samples. This opens MS-based insights in areas where methods like proximity labeling cannot operate:
- Human samples
- Archived tissue
- Cells that are difficult for transfection
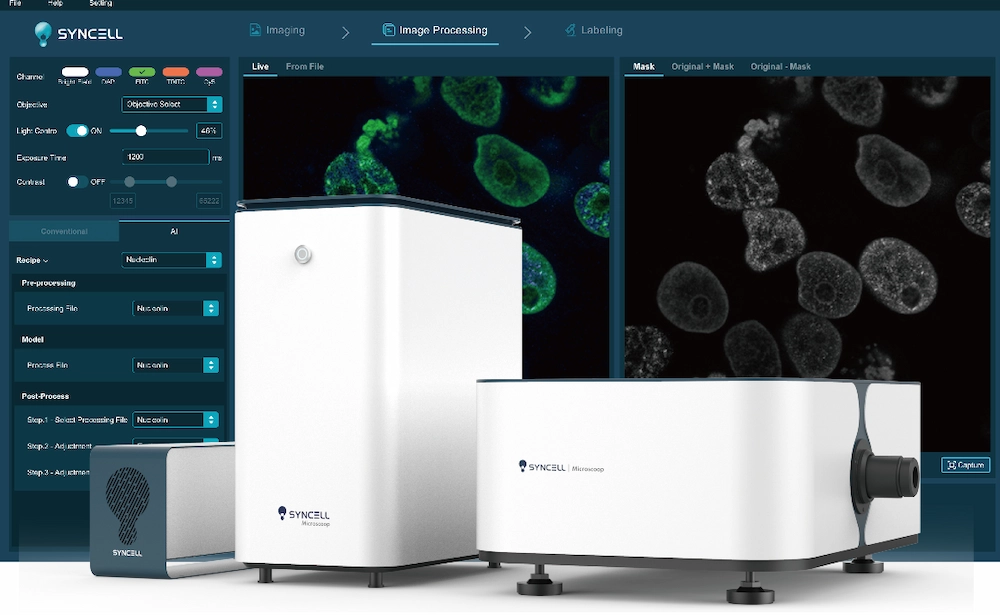
Spatial Proteomics Advantage
See Biology Like Never Before
Subcellular Resolution
Our advanced technology enables precise localization of proteins within cellular compartments, revealing intricate molecular interactions and pathways.

Unbiased Discovery
Explore the proteome without preconceived hypotheses, identifying novel biomarkers and therapeutic targets through comprehensive spatial analysis.
Microscopy-Guided Analysis
Integrate high-resolution imaging with proteomic data to correlate morphological features with molecular expression patterns.
Automated Workflows
Streamline your research with our end-to-end automated solutions, from sample preparation to data analysis and visualization.
Our Multi-Step Discovery Workflow
This streamlined process is designed to deliver comprehensive and actionable data from complex biological samples, ensuring high quality and resolution at every stage.
 >
>
Step 1: Sample Preparation & Acquisition
Precise tissue handling and preparation, followed by high-throughput data acquisition, ensures the integrity of the proteome for downstream analysis.
 >
>
Step 2: Data Processing & Alignment
Raw data is rigorously filtered, normalized, and aligned using proprietary algorithms to maximize feature detection and ensure accurate quantification.
 >
>
Step 3: Proteomic Analysis & Quantification
In-depth statistical analysis is performed to identify differentially expressed proteins and quantify their abundance across various sample cohorts.
 >
>
Step 4: Biological Interpretation & Reporting
The resulting protein lists are mapped to functional pathways and processes, generating a clear, comprehensive report for actionable biological insights.
Key Analytical Advantages
High-throughput analysis across multiple cells and tissue regions.
Real-time visualization and protein mapping.
Compatibility with advanced proteomics workflows.
Comprehensive integration of spatial and molecular data.
Performance Highlights
Powerful Performance in an Entry-Level System
Microscoop® — Illuminating the Hidden World of Spatial Proteomics
Microscoop® redefines proteomics by merging microscopy and protein discovery into a single platform. It allows scientists to uncover cellular and molecular complexity at a depth never before possible, driving breakthroughs in disease research, drug discovery, and systems biology.
Research Citation
| No. | Website | Category | Citation |
|---|

