Genomics


AVITI 24
Accelerate Your Discovery
Powered by the flexibility of ABC sequencing, AVITI24 is the first and only platform to enable co-detection of RNA, protein, and morphology and NGS in one dual-sided run.

AVITI
Innovative Next-Generation Sequencing
AVITI is a benchtop sequencing instrument with reimagined core technology to deliver flexibility and affordability while setting the standard for data quality.

AVITI LT
Budget-Friendly Sequencing Solution
Offering the same technology and features as AVITI, AVITI LT provides an ABC sequencing solution to suit your budget. The only difference is throughput.
Element Biosciences - Chemistries, Technologies and Workflows
- PCR-free amplification: Unlike other NGS platforms that use PCR-based bridge amplification, ABC uses rolling circle amplification (RCA). A circularized DNA library is used as a template to generate a continuous strand of DNA that forms a “polony”—a cluster of identical library copies.
- Separate sequencing steps: Traditional SBS combines base detection and strand extension into a single enzymatic step, which requires high concentrations of reagents. ABC separates these steps to optimize conditions for each.
- Avidite-based detection: Instead of using nucleotides with a reversible terminator and cleavable fluorophore, ABC uses fluorescently labeled “avidites”. These are polymers carrying many identical nucleotides that bind to multiple sites on the polony. Their strong, multivalent binding creates an ultrastable complex for imaging, allowing for accurate base detection with a 100-fold lower reagent concentration than SBS. After imaging, the avidites are washed away without damaging the DNA, leaving it “scarless” for the next cycle.
Avidite Based Chemistry (ABC) sequencing

Key Features
- Rolling circle amplification (RCA): This PCR-free amplification method reduces common sequencing artifacts such as index hopping, optical duplicates, and errors in difficult-to-sequence regions like homopolymers.
- Low-binding surface chemistry: The flow cell is coated with a low-binding surface chemistry that minimizes non-specific binding. This increases the signal-to-noise ratio, leading to clearer images and higher accuracy, even at high polony densities.
- Two-step sequencing: By decoupling base detection from strand extension, ABC can optimize the enzyme and reagent conditions for each step individually. This increases accuracy and efficiency while significantly reducing reagent usage.
- Modular design: The underlying ABC technology is flexible and extensible. Features like the polymer core, dyes, and nucleotide linkers can be optimized for different applications, from standard DNA and RNA sequencing to multiomic workflows.
- Broad library compatibility: ABC supports a wide range of sequencing applications and is compatible with both native Element libraries and most third-party libraries.
Advantages
High accuracy: ABC’s combination of RCA, optimized enzymatic steps, and multivalent avidite binding leads to superior data quality. Studies have shown higher mapping and variant calling accuracy, especially in challenging regions like homopolymers, when compared to other platforms.
Lower sequencing costs: The high stability of the avidite complexes enables the use of nanomolar concentrations of reagents, a massive reduction compared to traditional SBS. This significantly lowers the cost per gigabase.
Increased flexibility: The dual, independent flow cell design of the AVITI systems, combined with ABC’s robust chemistry, allows for flexible throughput and the ability to run two different experiments simultaneously. This reduces the need for batching and improves lab efficiency.
Reduced artifacts: The PCR-free RCA eliminates amplification bias and error propagation that can affect PCR-based methods, resulting in higher library complexity and lower duplication rates.
Cloudbreak sequencing and Teton CytoProfiling

AVITI24 - one instrument to replace multiple instruments for true 5D multiomics applications

Element Biosciences’ Cloudbreak and Cloudbreak Freestyle are kits for the AVITI System that employ avidite base chemistry (ABC) to provide fast, high-accuracy sequencing. The primary difference lies in the workflow and library compatibility: Cloudbreak Freestyle simplifies the process by performing a key step on-instrument, while the original Cloudbreak requires more manual preparation.
Cloudbreak sequencing
The standard Cloudbreak workflow requires that libraries be circularized on the benchtop, before being loaded onto the AVITI System.
- Circularization: This step, which is necessary for Element’s sequencing chemistry, must be performed manually by the user, outside of the instrument.
- Library compatibility: The original Cloudbreak chemistry is compatible with Element’s Elevate libraries and some third-party libraries, but other third-party libraries may require special handling or additional steps.
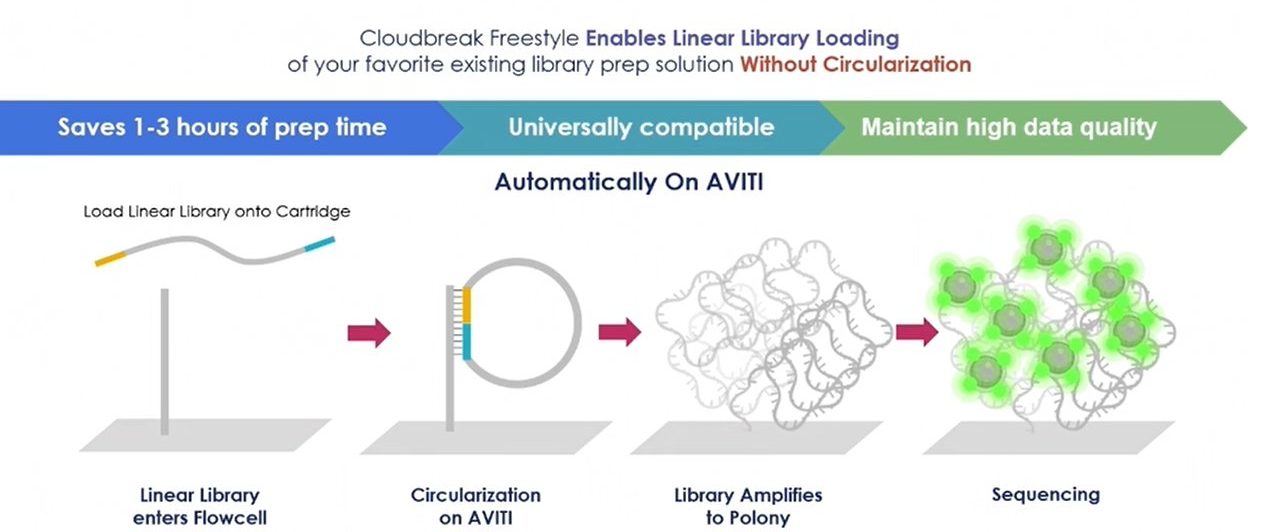
Cloudbreak Freestyle sequencing
Cloudbreak Freestyle represents an evolution of the Cloudbreak technology that removes manual steps and increases compatibility.
- Integrated circularization: The AVITI System performs the circularization step automatically on the flow cell as part of the run.
- Simplified workflow: Because circularization is automated, users can load linear libraries directly onto the instrument, saving preparation time and reducing hands-on steps.
- Enhanced compatibility: Cloudbreak Freestyle significantly expands support for various library types, including most third-party libraries, without the need for manual conversion. It can accommodate libraries with minor modifications or truncations to the adapter ends that would have caused issues with the original Cloudbreak chemistry.
Expert mode HD (High-Output mode)
This is not a separate kit, but a feature enabled on the AVITI System that boosts performance for all Cloudbreak sequencing kits.
- Higher output: Enables users to generate a greater number of reads per run, which can be advantageous for high-throughput applications like single-cell sequencing.
- Reduced cost: The increased output can lower the cost-per-gigabase by more than 30%.
- Software option: This is an instrument software setting, not a specific kit.
Comparison of Cloudbreak and Cloudbreak Freestyle (+ High-Output mode)
| Feature | Cloudbreak | Cloudbreak Freestyle | Cloudbreak Freestyle + High-Output Mode |
|---|---|---|---|
| Library Format | Requires libraries to be circularized before loading. | Directly accepts linear libraries for sequencing. | Same as Cloudbreak Freestyle, accepting linear libraries. |
| Workflow | Requires manual, benchtop library conversion. | Automated, on-instrument circularization, eliminating manual steps. | Same streamlined workflow as Cloudbreak Freestyle. |
| Library Compatibility | Standard Elevate, Nextera, and TruSeq libraries. | Enhanced compatibility with most third-party libraries and some small RNA adapters. | Same broad compatibility as Cloudbreak Freestyle. |
| Output | Standard output. | Standard output. | Up to 70% more reads per run, depending on the kit and settings. |
| Cost | Standard cost-per-gigabase. | Standard cost-per-gigabase. | Significantly reduced cost-per-gigabase due to higher output. |
| Configuration | Depends on the selected kit (e.g., read length). | Depends on the selected kit (e.g., read length). | Software-enabled option available on the AVITI and AVITI24 Systems. |
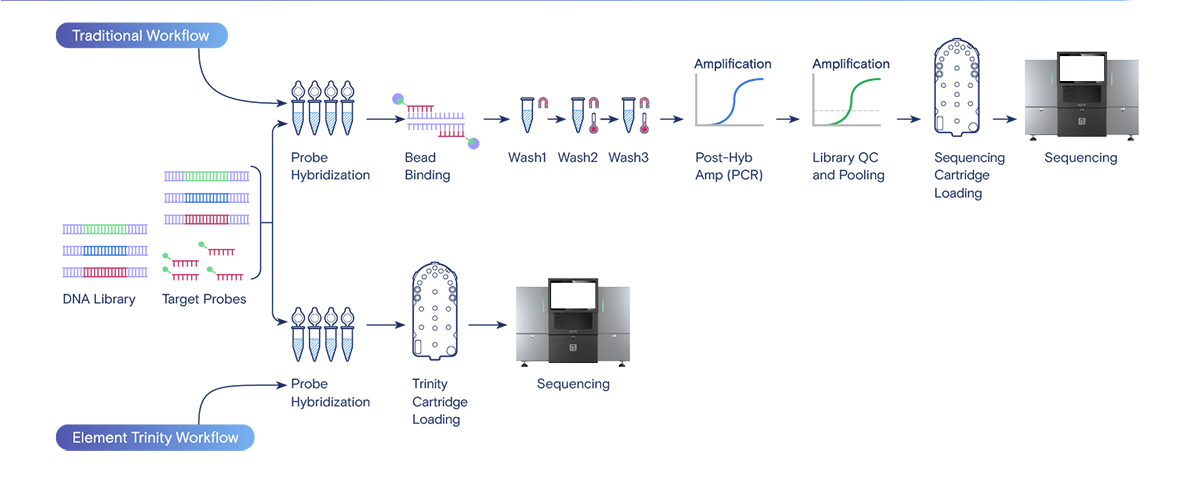
Trinity workflow is a novel hybrid capture solution for targeted sequencing that automates and streamlines traditionally complex, manual steps directly on the AVITI sequencer. By moving the enrichment steps from the benchtop to the instrument, it significantly reduces hands-on time, decreases turnaround time, and improves data quality.
Key features of the Trinity workflow
- Integrated capture and enrichment: Trinity utilizes a new flow cell and paired reagents that capture the target library fragments directly on the instrument after an initial hybridization step. This eliminates the need for manual, time-consuming steps involving magnetic beads, washes, and centrifugation.
- Reduced turnaround time: By automating the post-hybridization steps, the Trinity workflow drastically shortens the total time required for targeted sequencing. Workflows that typically took multiple days can now be completed in as little as 5 hours with a 1-hour fast hybridization option.
- Improved data quality: The elimination of post-capture PCR amplification, a standard step in traditional hybrid capture, leads to higher library complexity, lower duplication rates, and better variant calling accuracy, especially for indels.
- PCR-free option: Since the AVITI system uses a rolling circle amplification (RCA) chemistry rather than PCR for library amplification, the Trinity workflow is compatible with an entirely PCR-free targeted sequencing workflow. This further enhances data quality by avoiding common PCR-induced errors.
- Broad panel compatibility: While initially launched for exome sequencing, the workflow is also compatible with a wide range of targeted panels, including smaller gene panels for cancer research. Element Biosciences has partnered with companies like Integrated DNA Technologies (IDT) and Twist Bioscience for compatible reagents.
Comparison between traditional and Element Trinity workflows for Target Capture and Whole Exome Sequencing
| Feature | Traditional Hybrid Capture | Trinity Workflow |
|---|---|---|
| Post-hybridization steps | Manual process involving bead binding, washes, and post-capture PCR amplification. | Integrated and automated on the AVITI system, eliminating manual steps. |
| Hands-on time | Labor-intensive, taking several hours of hands-on time over two or more days. | Significantly reduced to as little as one hour of hands-on time for the entire capture workflow. |
| Turnaround time | Multiple days to complete the library preparation, enrichment, and sequencing setup. | As fast as a single day from library to data, with a 1-hour fast hybridization option. |
| Data quality | Prone to lower library complexity and higher duplication rates due to post-capture PCR. | Features higher library complexity, lower duplication rates, and improved variant calling accuracy. |
| Equipment | Requires a thermocycler, magnets, and other benchtop equipment for the wash steps. | Integrates the capture process into the AVITI system, eliminating the need for specialized external equipment. |
Advantages
- Preservation of spatial context: By sequencing directly within the intact cell, DISS preserves the spatial and molecular context of the sample, which is lost in traditional methods that require sample disruption.
- Reduced hands-on time: The automated, library-prep free process drastically reduces the manual effort and complexity of multiomic experiments, simplifying the entire workflow.
- Faster time to results: The streamlined workflow delivers high-resolution insights in days, as opposed to the weeks often required by multi-system, multi-step protocols.
- High data quality: The direct sequencing approach and integrated analysis improve data accuracy and reduce PCR-induced artifacts, leading to higher-confidence results.
- Flexibility and new applications: The customizable nature of DISS opens the door to new applications, such as unbiased cell typing, CRISPR screening, and multiomic functional genomics, all within a single automated system.
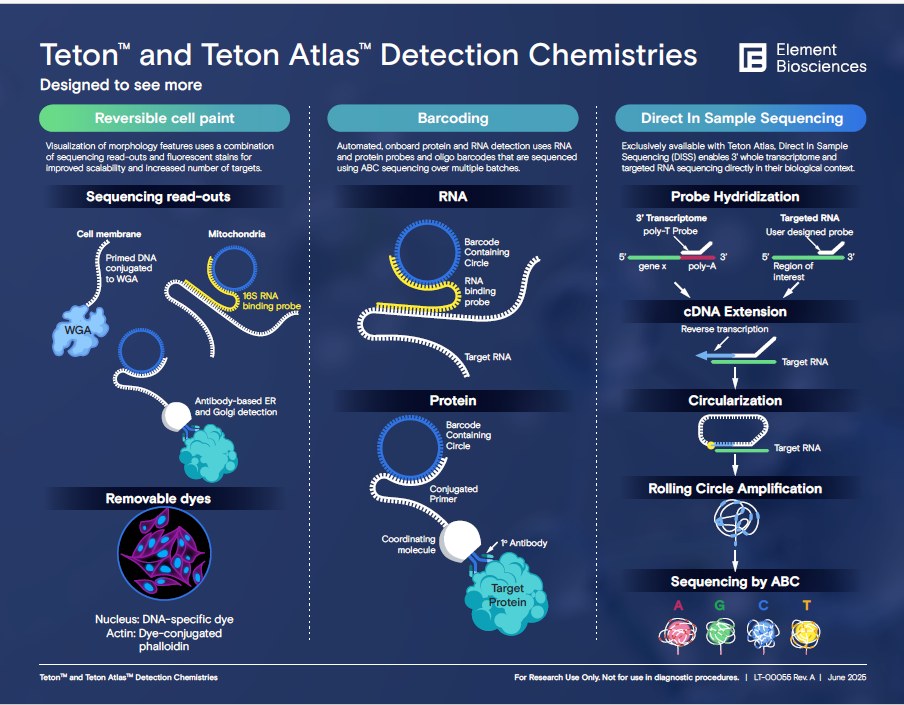
Element Biosciences’ Teton is a technology, featured on the AVITI24 System, that uses avidite base chemistry (ABC) and high-resolution imaging to enable Teton Cytoprofiling. Teton Cytoprofiling is a spatial multiomics method for analyzing RNA, protein, and cell morphology within a single experiment at single-cell resolution.
Teton
The Teton name refers to the core technology that enables the multiomic capabilities on the AVITI24 System. It is the foundation for performing cytoprofiling experiments and consists of specialized onboard chemistries and detection probes.
Key components of the Teton technology include:
- Probes and antibodies: A variety of detection probes and antibodies are used to label specific RNA, protein, and morphology markers within the cells.
- Avidite base chemistry (ABC): This proprietary chemistry is adapted from Element’s sequencing platform to provide high-resolution imaging and detection of the labeled probes.
- Onboard analysis: The AVITI24 instrument uses machine learning models to perform automated cell segmentation and primary data analysis, accurately assigning molecular features to individual cells.
Teton Cytoprofiling
Teton Cytoprofiling is designed to provide comprehensive, in-situ multiomic analysis directly on cells fixed to a flow cell, without the need for extensive library preparation. This integrated approach provides a complete view of cell biology in a single 24-hour run.
Key features of Teton Cytoprofiling include:
- Comprehensive multiomics: It enables the simultaneous visualization and analysis of RNA, proteins, and cell morphology. This allows researchers to understand complex biological pathways and the interplay between these different molecular features.
- Subcellular spatial resolution: By imaging intact cells on the flow cell, Teton can pinpoint the location of RNA and protein expression within each individual cell.
- High throughput: The AVITI24 System with Teton can profile up to 2 million cells in a single run, making it suitable for large-scale studies and screening.
- Simplified workflow: With less than one hour of hands-on sample preparation, the workflow is significantly more streamlined compared to traditional multiomic approaches.
- Flexible panels: Teton offers both pre-designed panels for specific research areas (e.g., immuno-oncology, neuroscience) and options for custom panels using the user’s own antibodies.
- Direct In-Sample Sequencing (DISS): Teton Atlas, a related chemistry, supports in-situ sequencing applications directly on the cells, enabling targeted or untargeted transcriptome analysis.
Teton Atlas is a multiomic technology from Element Biosciences that enables Direct In Sample Sequencing (DISS) on the AVITI24 System. It is an evolution of the Teton Cytoprofiling platform, expanding its capabilities from targeted RNA counting to direct sequencing of RNA, protein, and morphology within intact cells, without requiring traditional library preparation. This eliminates extensive manual work and preserves the native biological context of the sample.
Key features
- Direct In Sample Sequencing (DISS): Teton Atlas’s core innovation is its ability to perform sequencing of RNA directly within cells. It captures both targeted RNA sequencing (100+ base pairs) and unbiased 3′ whole-transcriptome data.
- 5D Multiomics: Teton Atlas can combine DISS with other Teton detection chemistries to measure up to five dimensions of cellular biology in a single experiment:
- RNA sequencing (targeted and whole-transcriptome)
- Protein expression (using custom or pre-designed panels)
- Phosphorylated protein detection
- Cell morphology (using reversible cell paints)
- Spatial location (subcellular resolution)
- Fully automated and flexible: The AVITI24 platform automates the DISS workflow from start to finish, from sample loading to primary analysis. Users can design custom workflows using an intuitive cloud-based designer to combine different multiomic readouts.
- Multiple configurations: The technology is available in different kit formats to suit various research needs. This includes a Low Output kit for targeted sequencing and CRISPR screening, as well as a High Output kit (expected later in 2025) for larger-scale discovery experiments.
- Species- and sample-type agnostic: Teton Atlas is compatible with a wide range of sample types, including human and non-human, primary cells, and cell lines.
| Teton Atlas Low Output (now available) | Teton Atlas High Output (coming in Q4 2025) | |
|---|---|---|
| Cell Paint | 3-plex | 6-plex |
| RNA counting | - | 3' WT or up to 500 Genes |
| RNA sequencing | Up to 100bp | Up to 100bp |
| Protein | Up to 88-plex | Up to 88-plex |
Advantages
- Streamlined workflow: By removing the need for labor-intensive library preparation, DISS significantly reduces hands-on time and complexity, freeing up researchers’ time and accelerating discovery.
- Preserved biological context: The in-situ sequencing approach maintains the spatial and molecular context of the sample. This allows for direct correlation of RNA, protein, and morphological features within individual cells.
- High data quality: Teton Atlas provides high-quality sequencing data (Q30+ for 100+ cycles) directly from cells. This offers high confidence for applications like unbiased cell typing, mutation detection, and CRISPR screening.
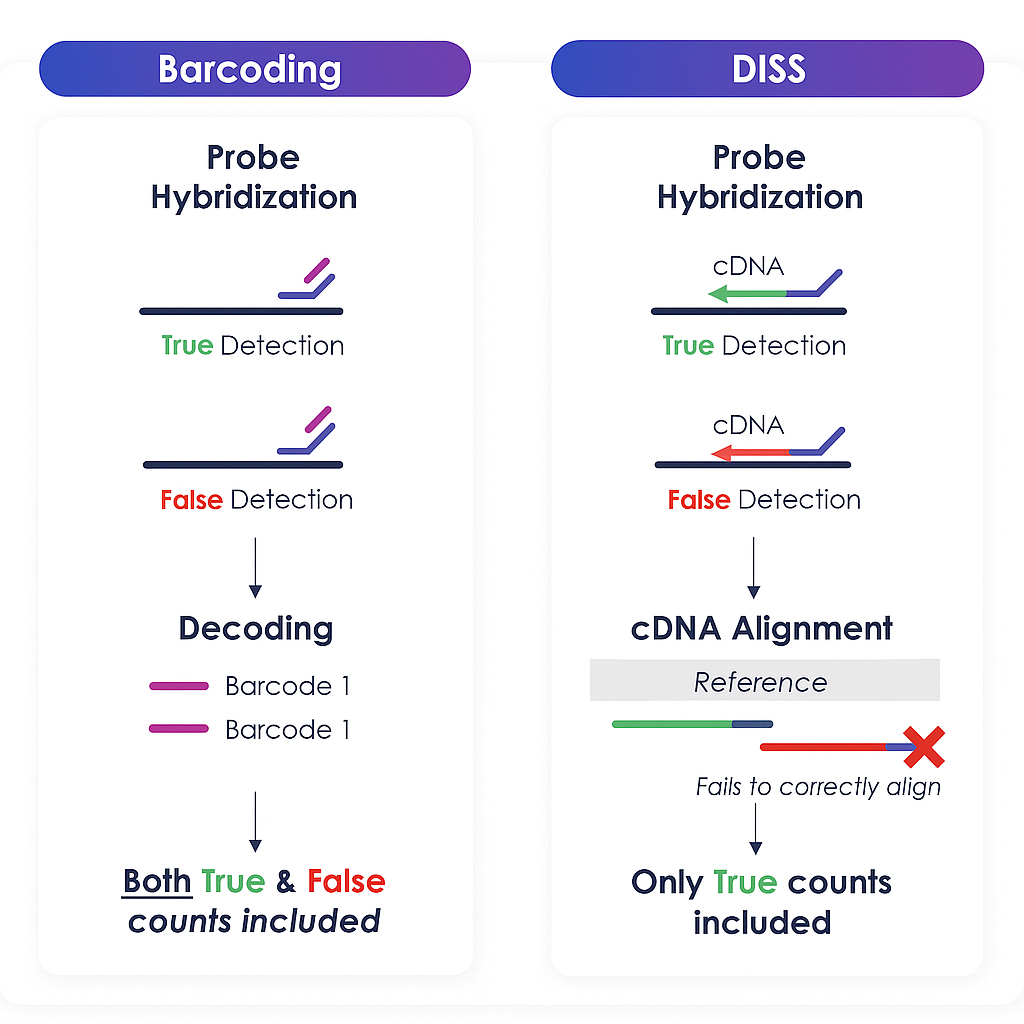
- Increased data resolution: Compared to probe-based counting methods, DISS offers higher specificity by providing the full sequencing information of the target gene, reducing the risk of off-target binding.
- Rapid insights: The fully automated, multiomic workflow enables comprehensive data acquisition in as little as 24 hours, dramatically accelerating the time from sample to actionable results.
- Enhanced flexibility: The modular design of Teton Atlas allows researchers to customize their experiments on-demand and adjust panels as their research evolves, rather than being restricted by rigid, pre-set protocols.
Cloudbreak sequencing and Teton CytoProfiling
AVITI24 - one instrument to replace multiple instruments for true 5D multiomics applications

Direct In Sample Sequencing (DISS) is a workflow on the Element Biosciences AVITI24 System that eliminates the need for traditional library preparation by directly sequencing nucleic acids within intact cells. As part of the Teton Atlas platform, DISS enables simultaneous multiomic profiling of RNA, proteins, and cell morphology at single-cell resolution.
Key features
- Integrated multiomics: DISS enables the acquisition of multiomic data (RNA, protein, and cell morphology) from the same individual cells in a single, automated run.
- Library-prep free: This is the core feature of the DISS workflow. By removing the need for manual library preparation, it saves significant hands-on time and reduces workflow complexity.
- High specificity: DISS directly sequences RNA within the cellular context and aligns the resulting reads. This provides a higher level of specificity compared to traditional probe-based methods, which can suffer from off-target binding and false positives.
- High-quality sequencing: The workflow is built on Element’s proven AVITI sequencing chemistry, which delivers high-quality data (Q30+) for reads up to 100 base pairs directly from the cells.
- Customizable and scalable: Researchers can tailor the workflow by designing custom assays, including targeted RNA panels and CRISPR screens. The AVITI24 platform allows for scalable experiments, from hypothesis-driven studies to large-scale discovery.
Direct In Sample Sequencing (DISS) with Onboard Detection Chemistry
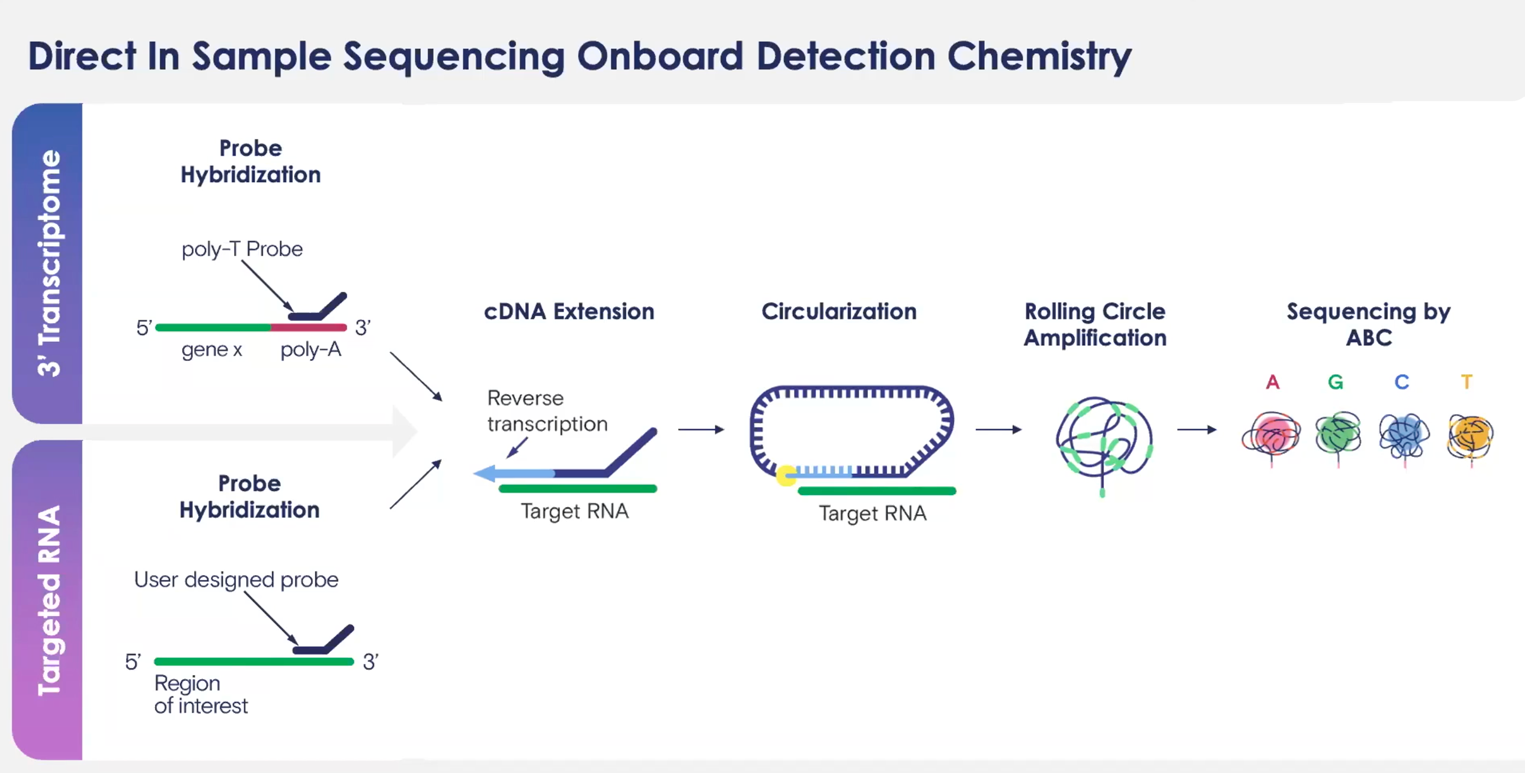
DISS overcomes Off-Target Binding vs Barcoding Approaches

N6 Tec is a company that develops advanced technology for the field of genomics, with a focus on improving the processes of DNA amplification and sequencing. Their main product is called iconPCR™

iconPCR
Accelerate Your Discovery
iconPCR™ is the first and only technology that intelligently adjusts amplification in real time (with AutoNorm) —so you get normalized, sequencing-ready libraries in one automated step, with no individual cleanup or extra reagents, while getting the best possible data from every sample.

ID3EAL Suite
Discovery Solutions
Exceptional miRNA detection powered by
the class-leading ID3EAL RT-qPCR technology

ID3EAL™ PanoramiR Knowledge Panel
Minimal Input, Maximum Insight
The ID3EAL™ PanoramiR Knowledge Panel is an effective approach for profiling miRNAs important in biological processes, disease mechanisms, or biomarker discovery for clinical applications such as disease diagnosis and prognosis, disease stratification or treatment selection.




